A massive problem in treating infectious diseases caused by bacteria is the development of bacterial strains that are increasingly resistant to antibiotics. The urgent question for doctors and drug companies is how to fight the emergence of resistant strains so that the known arsenal of antibiotics will remain effective. To address this problem, researchers have been working to understand how bacteria become resistant in the first place. Some new research published last week in Nature provides some major clues about surprising tricks bacteria use to combat antibiotics. (Nature 2010, 467, 82-85)
It turns out that just a few highly resistant bacteria can protect an entire population of bacteria from the effects of an antibiotic, even if most of the bacteria don’t have any inherent resistance. This remarkable conclusion was reached after a group of scientists conducted a series of elegant experiments using a strain of E. coli.
The researchers grew a bacterial culture from a strain of E. coli that can be killed by the antibiotic norfloxacin (that is, a non-resistant strain). They then subjected the bacteria to a dose of the antibiotic that was strong enough to stress the culture (inhibit growth by about 60%) but too weak to actually kill the bacteria off. For the first couple of days, the bacteria growth was effectively slowed, but then they seemed to become more resistant and started growing faster on the third day. So the researchers upped the dose of antibiotic, making it strong enough to again inhibit growth by about 60%. The same thing happened – growth was slowed at first, but then picked up again. After a few iterations of this back-and-forth, the bacteria population was able to tolerate quite a high dose of antibiotic. This emergence of antibiotic resistance has been seen many times before; however, the surprising result is what the researchers found when they looked at individual bacteria cells throughout the 10-day study.
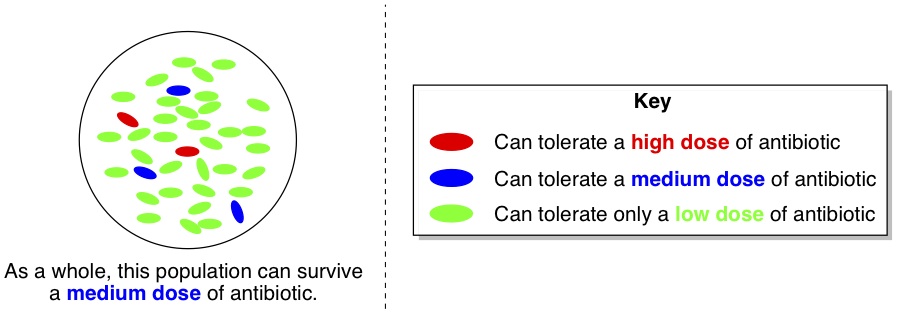
Each day, the researchers isolated a dozen or so individual bacteria cells and tested their resistance to the antibiotic. Now, let’s say that the whole population of bacteria was able to stand a “medium” dose of antibiotic. One would expect that cultures grown from randomly chosen individual cells would have the same degree of tolerance, and thus could also stand a “medium” dose of antibiotic. Curiously, this wasn’t the case. Almost all of the isolated cells actually had much lower tolerance (green cells, below) to the antibiotic than the population as a whole. However, a very small minority of the cells (red ones) had higher tolerance than the whole population.
So how was the population of bacteria surviving a “medium” dose of antibiotic, when almost all of the cells only had a “low” level of tolerance? To answer this question, the researchers analyzed the proteins and other byproducts that were being produced by the high-resistance (red) and the low-resistance (green) bacteria in the presence of the antibiotic. They observed a trend – although a particular protein called TnaA was being produced like normal by the high-resistance bacteria, this protein was suspiciously absent in the material analyzed from the low-resistance bacteria. The protein TnaA is responsible for making a molecule called indole.
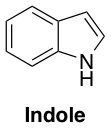 This was intriguing, because indole has been implicated as something that helps protect E. coli from damage. When the bacteria weren’t stressed by antibiotics, both the high-resistance and the low-resistance isolated bacteria produced the same amount of indole, but under antibiotic stress, the low-resistance bacteria lost their ability to make indole because they weren’t making TnaA anymore. However, when the researchers helped them out by adding some extra indole to the low-resistance bacteria, these bacteria became able to tolerate a “medium” dose of antibiotic instead of just a low dose. Presumably, the indole was providing some damage protection for the stressed bacteria.
This was intriguing, because indole has been implicated as something that helps protect E. coli from damage. When the bacteria weren’t stressed by antibiotics, both the high-resistance and the low-resistance isolated bacteria produced the same amount of indole, but under antibiotic stress, the low-resistance bacteria lost their ability to make indole because they weren’t making TnaA anymore. However, when the researchers helped them out by adding some extra indole to the low-resistance bacteria, these bacteria became able to tolerate a “medium” dose of antibiotic instead of just a low dose. Presumably, the indole was providing some damage protection for the stressed bacteria.
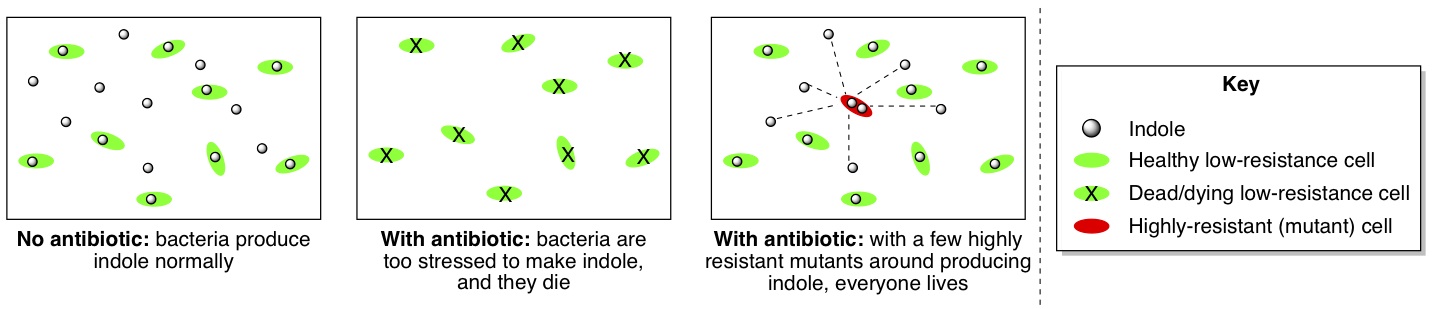
These results led the researchers to conclude that in the whole population of bacteria undergoing the 10-day antibiotic stress test, the few high-resistance bacteria were actually helping all the other bacteria by producing indole for everyone to share (below, third picture). Even though the majority of the bacteria couldn’t produce their own indole when they were stressed by antibiotic, the indole produced by the few high-resistance bacteria was protecting everyone else from damage. For this reason, the population as a whole could withstand a medium dose of antibiotic, even though most of the individual cells couldn’t stand more than a low dose on their own.
Furthermore, the researchers found that making indole carries a “fitness cost” for bacteria – they can’t grow as fast when they’re having to make indole. Thus, when the high-resistance bacteria produce indole for the whole group, they are actually sacrificing their own ability to grow quickly for the sake of the entire population’s ability to grow at all. How charitable!
Nevertheless, despite how generous these bacteria appear to be, the ultimate goal is to figure out how to kill them. Up until now, most research has focused on how genetic mutations arise and enable bacteria to become resistant to antibiotics. However, this publication demonstrates that there is more going on than just mutations in individual bacteria. The dynamics of a bacterial community as a whole also plays a role in antibiotic resistance. Future research is likely to uncover more ways that bacteria communicate with each other and, hopefully, provide some insight into new ways to combat the emergence of antibiotic resistance in harmful bacteria.